Redox proteins
Nature provides a wealth of enzymes that link the two half reactions of a redox reaction through an electron transfer chain. Biological processes, such as respiration, anaerobic metabolism, and nitrogen fixation, depend on these enzymes and offer inspirational alternatives for difficult chemical conversions, via redox proteins.
Format dehydrogenase
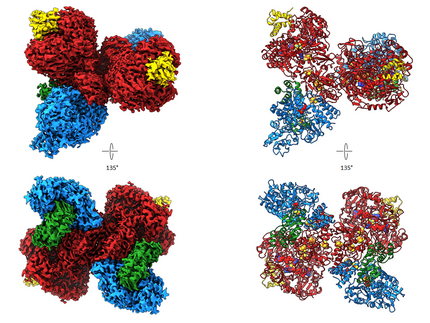
In 2020, we published the first cryo EM structure of a molybdoenzyme, the redox protein formats dehydrogenase from Rhodobacter capsulatus (Rc FDH ) . This enzyme catalyses the reversible oxidation of formats to carbon dioxide and is an interesting target for biotechnological applications such as carbon sequestration or hydrogen storage. We presented the 3.3 Å cryo-EM structures of Rc FDH as isolated and in the presence of NADH. The structures reveal a complex arrangement of Fe-S clusters in the dimer, a conserved binding mode of the FdsD to the FdsA subunit and that NADH reduction leads to charging of electron carrying cofactors of RcFDH.
The purified complex consists of four subunits, FdsA, FdsB, FdsG and FdsD and forms a 360 kDa dimer of FdsABGD heterotetramers. The heterotetramers adopt an elongated structure with dimensions of 140 × 80 × 77Å and are arranged in an almost perpendicular back-to-back orientation in the dimer. Subunit FdsA harbors the bis-metal-binding pterin guanine dinucleotide cofactor that binds the molybdenum and creates the oxygen sensitive active site. Electrons gained from oxidation formats are transferred via a 76 Å long electron transfer chain consisting of four [4Fe-4S] clusters and one [2Fe-2S] clusters onto FMN, and finally NAD +. The electron transfer chains in the protomers of the dimer are connected by two [4Fe-4S] clusters located at the dimer interface, allowing for electron exchange between the two protomers. When RcFDH is reduced with NADH + H +, electron carrying cofactors show scattering behavior that can only be explained by a change of charge in comparison with the as isolated enzyme, suggesting that comparative cryo-EM can be used to visualize charge differences in different states of redox enzymes.
Photosystem I
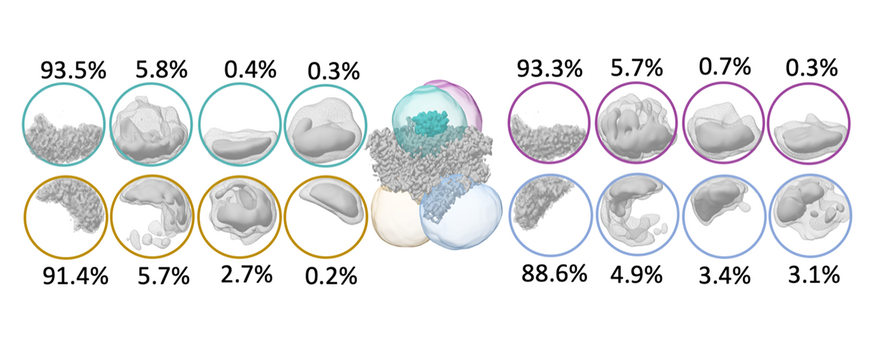
Photosystem I converts solar energy into electrical energy by oxidizing the soluble redox mediator cytochrome c6 and reducing ferredoxin. To characterize the transient binding of cytochrome c6 to Thermosynechoccus elongatus photosystem I the cryo EM structure of the cross-linked complex was solved to 2.9 Å resolution in collaboration with UniSysCat member Athina Zouni from Humboldt University and Fred Lisdat from TU Wildau.
We were able to identify additional cofactors and side chain density of the subunit PsaK. Despite SANS data indicate a complex formation between TePSI and non-native cytochrome from horse heart we could not identify Cyt c6 in our cryo-EM analysis due to the poor binding specificity. These results reveal the difficulty to identify very small binding partners in cryo EM structures with low binding affinities.
Publications:
Radon C., Mittelstädt G., Duffus BR, Bürger J, Hartmann T., Mielke T, Teutloff C, Leimkühler S, Wendler P (2020) Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase. Nat. Commun. 11:1912. doi: 10.1038/s41467-020-15614-0
Kölsch A., C. Radon, M. Golub, A. Baumert, J. Bürger, T. Mielke, F. Lisdat, A. Feoktystov, J. Pieper, A. Zouni, P. Wendler (2020) Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I. Cur Res in Struc Biol 2:171-179. doi: 10.1016/ j.crstbi.2020.08.003