Peroxisomal Protein Import
Peroxisomes are single membrane cell organelles that contain at least 50 different enzymes involved in essential metabolic pathways including the oxidative breakdown of long-chain fatty acids, peroxide detoxification, and the synthesis of etherphospholipids. They are abundant in the liver, kidney and central nervous system and can adopt their number and size dramatically in individual cell or tissue types according to environmental cues. Peroxisomal membrane transporters have to actively navigate metabolites, such as electron carriers (e.g. NAD/H+) and nucleotides (ATP) or even fully folded proteins across the peroxisomal membrane. At least 33 Pex proteins, so called peroxins, are known to participate in the formation and maintenance of peroxisomes in yeast. Mutations in one or more of the underlying PEX genes cause a broad spectrum of human diseases that are referred to as peroxisome biogenesis disorders (PBDs). The majority of peroxisomal matrix proteins contain a C-terminal peroxisomal targeting sequence type 1 (PTS1), which is recognised by the soluble receptor protein Pex5. The cargo-Pex5 complex is delivered to the peroxisomal membrane and is proposed to form a temporary protein conducting channel in combination with the peroxisomal membrane protein Pex14. PTS1 import depends on monoubiquitination and recovery of Pex5 from the peroxisomal membrane presumably to maintain the import cycle. This recycling step is ATP dependent and is carried out by the two type II AAA+ (ATPases associated with various cellular activities) peroxins Pex1 and Pex6. Type II ATPases contain two ATPase domains in tandem and their active state is usually a hexamer. Pex1 and Pex6 form a 700 kDa, heterohexameric complex, which is proposed to remodel its substrate by threading it through the central pore.
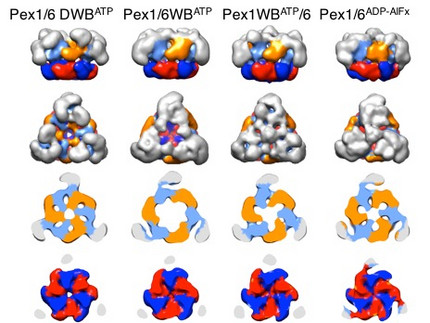
We solved low-resolution structures of yeast Pex1/6 complexes in 7 different states of the ATPase cycle, offering an exhaustive structural analysis of its nucleotide dependent dynamic movements. The complexes are trimers of Pex1/6 dimers with a distinct triangular shape. The ATPase activity of the complex is solely attributed to the second AAA+ domain (D2) of Pex6, whereas the D2 domains of Pex1 and Pex6 harbour substrate interaction loops essential for complex function in vivo. Our maps indicate that prolonged ATP binding to the Pex1/6 D2 domains results in a planar, hexameric arrangement of the D2 ring. However, nucleotide hydrolysis causes a downward rotation of the D2 domains. We propose that ATP hydrolysis can only occur, when interacting motifs such as the arginine finger of the neighbouring domain to the left, are positioned correctly to promote communication.
Publications:
- Ciniawsky, S., I. Grimm, D. Saffian, W. Girzalsky, R. Erdmann, P. Wendler (2015). Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat Commun. 6:7331. doi:10.1038/ncomms8331 https://www.nature.com/articles/ncomms8331
- Saffert, P., C. Enenkel, P. Wendler (2017). Structure and function of p97 and Pex1/6 Type II AAA+ complexes. Front. Mol. Biosci. 4:33. doi: 10.3389/fmolb.2017.00033 http://journal.frontiersin.org/article/10.3389/fmolb.2017.00033/full
- Schieferdecker A., P. Wendler (2019). Structural mapping of missense mutations in the Pex1/Pex6 complex. International Journal of Molecular Sciences. 20(15), 3756; https://doi.org/10.3390/ijms20153756