Molecular mechanisms of Hepatitis C virus (HCV) assembly
Hepatitis C Virus (HCV) constitutes a significant health burden worldwide, primarily due to the unavailability of specific vaccines as well as to the unaffordability of the existing therapies. Despite scientific advances in the field of HCV pathology, the development of a specifically aimed therapeutic approach is being challenged by the lack of fundamental knowledge of the molecular mechanisms involved in HCV life cycle within the infected host cells.
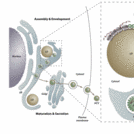
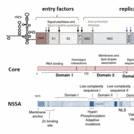
In this context, we propose to study two HCV proteins which are fundamental for virus formation: the core protein and NS5A. The HCV core protein is the first protein to be translated from the genome and forms the nucleocapsid. NS5A is a multifunctional phosphoprotein and essential for the HCV RNA replication, including the formation of a replication complex on endoplasmic reticulum (ER) membranes or lipid droplets and interaction with the core protein.
We will quantitatively study the interaction of these proteins with the host cell lipids by using Surface Plasmon Resonance (SPR) and in vitro model membrane systems mimicking lipid droplets (LDs) and the endoplasmic reticulum (ER), i.e. the cellular organelles where virus assembly occurs. In addition, protein-protein interactions driving core protein oligomerization – a crucial step in the assembly of new virions – will also be investigated quantitatively in living cells, by applying state-of-the-art microscopy methods, such as Number & Brightness microscopy (N&B), Spatial Intensity Distribution Analysis (SpIDA) and super-resolution microscopy. By combining the information deriving from model and cellular systems, we will be able to clarify the molecular mechanisms driving HCV capsid formation. Ultimately, this knowledge might be used to design specific antiviral approaches aiming at hindering virus replication and the spreading of infection.