Research topics
Overview
The microtubule cytoskeleton is of crucial importance for cellular architecture, transport and cell division, since it provides the tracks for organelle positioning and vesicle transport. During cell division, microtubules form the mitotic spindle, which is required for proper segregation of sister chromatid. The centrosome serves as the main microtubule organizing center and is bound to the cytosolic face of the nucleus.
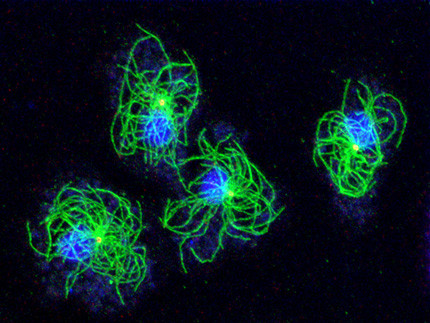
Our research group uses Dictyostelium amoebae (www.dictybase.org) as a model for the cell biology of the centrosome and the nuclear envelope. Similar to amoeboid animal cells such as neutrophils these amoebae contain a radial array of microtubules emanating from the centrosome. Unlike animal cells Dictyostelium centrosomes contain no centrioles but consist of a three-layered core structure surrounded by a corona harboring microtubule nucleation complexes. They duplicate in prophase of a semi-closed mitosis and organize a central spindle involved in centrosome separation and chromosome segregation.
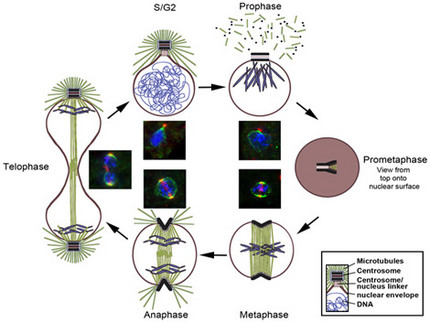
The Dictyostelium centrosome cycle
Dictyostelium centrosome cycle. Nuclei, chromosomes and centrosomes with attached mictrotubules are shown in schematic cross sections of different cell cycle stages, except for prometaphase where a view to the nuclear surface is depicted. Corresponding immunofluorescence microscopy images show staining of centrosomes (red), nuclear envelope and centrosome/nucleus linkage (green) and DNA (blue).
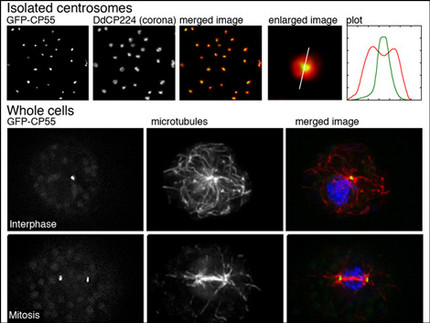
Functional analyses of centrosomes
We have established a procedure for preparative isolation of Dictyostelium centrosomes and have used these centrosomes for mass spectrometrical analysis of their protein composition. Thus we have identified 34 new centrosomal candidate proteins. Currently we functionally analyze these proteins using RNAi-strains, GFP-strains or knockout mutants. We employ modern microscopic methods and strategies to reveal protein-protein interactions with other known and unknown centrosomal components. A few example for the CP55 are shown in the image. Quite recently we have also established the Expansion Microscopy (ExM) superresolution method in Dictyostelium amoebae.
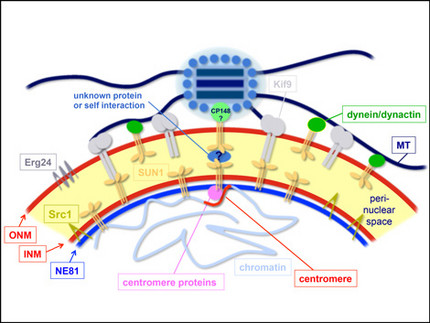
Centrosome/nucleus attachment and nuclear envelope organization
Centrosomal attachment to nuclei is crucial for proper mitosis and nuclear positioning in various organisms, and generally involves SUN-family proteins located at the inner nuclear envelope. There is still no common scheme for the outer nuclear membrane proteins interacting with SUN1 in centrosome/nucleus attachment. Our model employs Sun1 in both nuclear membranes and interacting motors such as dynein and the Kif9 kinesin.
The nuclear envelope (NE) consists of the outer and inner nuclear membrane, whereby the latter is bound to the nuclear lamina. With Dictyostelium NE81 we have identified the first bona fide lamin in a non-metazoan organism. Though proximity-dependent biotin identification (BioID) we could identify two conserved protein interactors at the inner nuclear membrane, i.e. Sun1 and Src1. Furthermore, by TEM, SEM and superresolution light microscopy we could show that NE81 assembles into filaments with a thickness in the range of intermediate filaments.
Methods used in the lab
1. Microscopic methods
These include live cell imaging of fluorescently labeled cells, fluorescence microscopy of fixed cells including superresolution techniques such as expansion microscopy (ExM), Airyscan microscopy and STED microscopy. Electron microscopy is performed in collaboration with Prof. Baumann (Dept of Animal Physiology UP) and Martin Goldberg (Durham, UK). We have full access to the advanced microscopy equipment of the institute including confocal laser scanning microscopes, TEM and live cell imaging setups such as wide-field deconvolution microscopes and a spinning disk confocal microscope.
2. Biochemical methods
Protein-protein interaction studies using BioID (proximity-dependent biotin identification), NanoTrap and/or TAP-tag affinity chromatography, co-immunoprecipitation. Protein separation and purification using electrophoresis, gradient centrifugation, gel filtration, ion exchange and affinity chromatography.
3. Molecular biology methods
DNA cloning, PCR, Dictyostelium transformation, knock-in and knock-out technologies, RNAi, two-hybrid analysis