Research
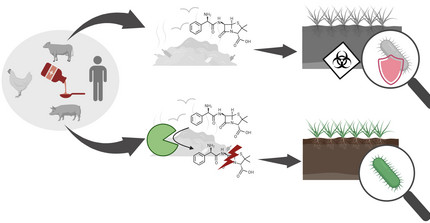
Antibiotics are an important milestone in medicine; however, their increased use has resulted in an alarming rise of antimicrobial resistances. Antibiotics are used in large quantities not only in human medicine but also in animal farming and agriculture, where they enter the environment through waste products, feces, or milk of treated cows, promoting antimicrobial resistance, and ultimately leading to untreatable multidrug-resistant pathogens threatening also human health. Importantly, antibiotic-resistant bacteria often activate export mechanisms that result in resistance to various structurally unrelated antibiotics. We are working on novel strategy for the enzymatic inactivation of antibiotics in different media. This facilitates, for example, antibiotic inactivation in whole milk, which can help to treat waste milk from medicated cows, to be fed to calves without the risk of inducing antibiotic resistances. Other applilcation areas we are looking into are the treatment of waste water, e.g. from hospitals or general waste water plants. We believe that our strategy can significantly reduce the amount of antibiotic in our environment, mitigating the occurrence and dissemination of resistances.
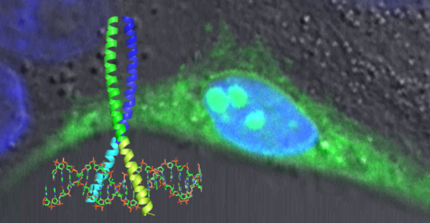
Protein-protein interactions are key for virtually all biological systems and play important roles in numerous diseases. Our research focuses on:
(i) understanding the molecular principles driving specificity and affinity of protein-protein interactions, and
(ii) generating inhibitors interfering with such protein interactions for analytical as well as medical purposes.
We use rational design in combination with in vivo and in vitro selection systems to generate such interfering peptides (iPEP). Remote control of inhibitor activity can be achieved by attaching photo-switchable chemical linkers. Quantitative data of inhibitor and wild-type interactions will aid modeling of the response of the underlying system. iPEPs are key tools for studying not only single pathways but also their interconnection on the systems level.
Our research combines synthetic aspects of biology with the system approach by generating synthetic peptide modules that are applicable for analyzing the integration of pathways in biological systems but can also be used to build artificial pathways to further our understanding of the system in general or validate hypotheses.